Abstract
Review Article
COVID-19 pandemic, recurrent outbreaks and prospects for assimilation of hCoV-19 into the human genome
Vinod Nikhra*
Published: 12 October, 2020 | Volume 4 - Issue 1 | Pages: 111-115
The outbreaks and resurgence: The disease which reportedly began in the Chinese city Wuhan in November-December 2019, soon spread to various parts of the world, and was named and declared a pandemic disease by WHO. While the European countries were recovering from the epidemic, the disease took hold in the USA, the South American countries, Arabian countries, and South Asian countries, predominantly affecting Brazil, Peru, Iran, and India. Presently, many European countries are witnessing a resurgence and recurrent outbreaks of COVID-19.
Spread and evolving new insights: Whereas there is workplace-related infection rise as people are returning to their offices, in other places the outbreaks are related to the people crowding and meeting care-freely and trying to resort back to their earlier way of life. The reopening of the educational facilities across the continents may make matters worse.
Impact on health and healthcare: Most cases of COVID-19 infections go unnoticed and are followed by self-recovery. But what may appear good from the clinical perspective, appears to complicate epidemiological efforts to contain the outbreak. With the evolving information about the disease, there seem to be certain possible outcomes such as control and containment, or the persistence of the disease as global endemic accompanied with outbreaks and resurgent episodes.
Gnetic factors linked to disease severity: With the COVID-19 pandemic, not all infected patients develop a severe respiratory illness. Further, there is a large variation in disease severity, which may be due to the genetic factors underlying the variable response to the virus. It is becoming clear that apart from the advanced age and pre-existing conditions, certain genetic constituent factors render some patients more vulnerable to the more severe forms of the diseases.
Integration of virus into human genome: A significant part of the human genome is derived from viruses especially the RNA viruses. In fact, about 8 percent of the human genome is made up of endogenous retroviruses (ERVs), which are viral gene sequences that have become a permanent part of the human lineage after they infected our ancient ancestors. With this background, a novel concept emerging that if COVID-19 persists for several generations, its genetic material is projected to be integrated or assimilated into human genome. The involved mechanisms are conceptualized through the transposons or transposable elements of the SARS-CoV-2.
Read Full Article HTML DOI: 10.29328/journal.ijcv.1001025 Cite this Article Read Full Article PDF
Keywords:
ACE-2 receptors; COVID-19 pandemic; Endogenous retroviruses (ERVs) ; Genetic factors; hCoV-19; Proofreading proteins; RNA viruses; SARSCoV-2; Transposons; VIPs
References
- Kupferschmidt K. Can Europe tame the pandemic's next wave? Science. 2020; 369: 1151-1152. PubMed: https://pubmed.ncbi.nlm.nih.gov/32883840/
- Tang S, Mao Y, Jones RM, Tan Q, Ji JS, et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ Int. 2020; 106039. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7413047/
- Kupferschmidt K. Case clustering emerges as key pandemic puzzle. Science. 2020; 368: 808-809. PubMed: https://pubmed.ncbi.nlm.nih.gov/32439769/
- Kupferschmidt K, Cohen J. Will novel virus go pandemic or be contained? Science. 2020; 367: 610-611. PubMed: https://pubmed.ncbi.nlm.nih.gov/32029604/
- Pollitt KJG, Peccia J, Ko AI, Kaminski N, Dela Cruz CS, et al. COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Human Genomics. 2020; 14; 17. PubMed: https://pubmed.ncbi.nlm.nih.gov/32398162/
- Murray MF, Kenny EE, Ritchie MD, Rader DJ, Bale AE, et al. COVID-19 outcomes and the human genome. Genet Med. 2020; 22: 1175–1177. PubMed: https://pubmed.ncbi.nlm.nih.gov/32393819/
- Wu BB, Gu DZ, Yu JN, Yang J, Shen WQ. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect Genet Evol. 2020; 84: 104485. PubMed: https://pubmed.ncbi.nlm.nih.gov/32739464/
- Nguyen A, David JK, Maden SK, et al. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J Virol. 2020; 94: e00510-20. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7307149/
- Moelling K, Broecker F. Viruses and Evolution - Viruses First? A Personal Perspective. Front Microbiol. 2019; 10: 523. PubMed: https://pubmed.ncbi.nlm.nih.gov/30941110/
- Callif BL. Organumics: An Epigenetic Re-framing of Life, Consciousness and Evolution. Kindle Edition, 2019 ASIN: B07ZMKMVSW, Publisher: S. Woodhouse Books, Everything Goes Media LLC.
- Tang W, Mun S, Joshi A, Han K, Liang P, et al. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res. 2018; 25: 5: 521-533. PubMed: https://pubmed.ncbi.nlm.nih.gov/30052927/
- Enard D, Petrov DA. Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. 2020.
- Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. ELife. 2016; 5: e12469. PubMed: https://pubmed.ncbi.nlm.nih.gov/27187613/
- Rosenberg NA, Kang TL. Genetic Diversity and Societally Important Disparities. Genetics. 2015; 201: 1–12. PubMed: https://pubmed.ncbi.nlm.nih.gov/26354973/
- Carrasco-Hernandez R, Jácome R, Vidal YL, de León SP. Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review. ILAR J. 2017; 58: 343–358. PubMed: https://pubmed.ncbi.nlm.nih.gov/28985316/
- Robson F, Khan KS, Le TK, et al. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Molecular Cell. 2020; 79: 710-727.
- Pérez-Losada M, Arenas M, Galáne JC, Bracho MA, Hillung J, et al. High-throughput sequencing (HTS) for the analysis of viral populations. Infect Genet Evol. 2020; 80: 104208. PubMed: https://pubmed.ncbi.nlm.nih.gov/32001386/
Figures:
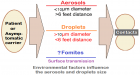
Figure 1

Figure 2

Figure 3
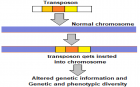
Figure 4
Similar Articles
-
Hypothesis about pathogenic action of Sars-COV-2Del Prete Salvatore*,Marasco Daniela,Sabetta Rosalaura. Hypothesis about pathogenic action of Sars-COV-2. . 2020 doi: 10.29328/journal.ijcv.1001009; 4: 021-022
-
The Psychology of the Common Cold and Influenza: Implications for COVID-19Andrew P Smith*. The Psychology of the Common Cold and Influenza: Implications for COVID-19. . 2020 doi: 10.29328/journal.ijcv.1001011; 4: 027-031
-
Exploring pathophysiology of COVID-19 infection: Faux espoir and dormant therapeutic optionsVinod Nikhra*. Exploring pathophysiology of COVID-19 infection: Faux espoir and dormant therapeutic options. . 2020 doi: 10.29328/journal.ijcv.1001013; 4: 034-040
-
Identifying patterns in COVID-19: Morbidity, recovery and the aftermathVinod Nikhra*. Identifying patterns in COVID-19: Morbidity, recovery and the aftermath. . 2020 doi: 10.29328/journal.ijcv.1001016; 4: 056-064
-
COVID-19 pandemic, recurrent outbreaks and prospects for assimilation of hCoV-19 into the human genomeVinod Nikhra*. COVID-19 pandemic, recurrent outbreaks and prospects for assimilation of hCoV-19 into the human genome. . 2020 doi: 10.29328/journal.ijcv.1001025; 4: 111-115
-
Stages in COVID-19 vaccine development: The Nemesis, the Hubris and the ElpisVinod Nikhra*. Stages in COVID-19 vaccine development: The Nemesis, the Hubris and the Elpis. . 2020 doi: 10.29328/journal.ijcv.1001028; 4: 126-135
-
Impact of COVID-19 pandemic on anti-microbial resistance and secondary microbial infectionsKP Mishra*,Priyanka Mishra,AK Singh,SB Singh. Impact of COVID-19 pandemic on anti-microbial resistance and secondary microbial infections. . 2021 doi: 10.29328/journal.ijcv.1001032; 5: 032-036
-
Viral overload of COVID-19 pandemics: Overweight people a soft target to get an infectionFaheem Anwar*,Muhammad Tayyab*,Ihteshamul Haq,Obaid Ullah Shah. Viral overload of COVID-19 pandemics: Overweight people a soft target to get an infection. . 2021 doi: 10.29328/journal.ijcv.1001037; 5: 070-071
-
On the effect of millimeter waves on DNA and RNA of virusesIkhlov BL*. On the effect of millimeter waves on DNA and RNA of viruses. . 2022 doi: 10.29328/journal.ijcv.1001046; 6: 029-033
-
Monkeypox virus outbreak: A new threat of virus to mankindSukhvir Kaur*. Monkeypox virus outbreak: A new threat of virus to mankind. . 2022 doi: 10.29328/journal.ijcv.1001048; 6: 038-042
Recently Viewed
-
Acyclovir Induced Acute Kidney Injury: A Case ReportZiauddin Mohammed*, Mariya Zoha Muskan, Megha Mohan Narayanan. Acyclovir Induced Acute Kidney Injury: A Case Report. Arch Pharm Pharma Sci. 2024: doi: 10.29328/journal.apps.1001048; 8: 001-002
-
Evaluation of the Anti-inflammatory Activity of Equisetum arvense and Baccharis trimera FractionsCarolina Ferreira Vaz, Alan Fernandes Mariano, Júlia Amanda Rodrigues Fracasso, Marcus Vinicius Vieitas Ramos, Lucineia dos Santos, Herbert Júnior Dias*. Evaluation of the Anti-inflammatory Activity of Equisetum arvense and Baccharis trimera Fractions. Arch Pharm Pharma Sci. 2024: doi: 10.29328/journal.apps.1001049; 8: 003-008
-
The Cortisol Connection: Weight Gain and Stress HormonesBalvinder Singh, Neelesh Kumar Maurya*. The Cortisol Connection: Weight Gain and Stress Hormones. Arch Pharm Pharma Sci. 2024: doi: 10.29328/journal.apps.1001050; 8: 009-013
-
Correlation of Inappropriate use of Ceftriaxone and Bacterial Resistance in the Hospital Environment: Integrative ReviewLarissa Furtado Abrantes, Joyce Lima de Sousa, Joel Messias Soares Ramos, Rafael Rodrigues Leite, Sávio Benvindo Ferreira*. Correlation of Inappropriate use of Ceftriaxone and Bacterial Resistance in the Hospital Environment: Integrative Review. Arch Pharm Pharma Sci. 2024: doi: 10.29328/journal.apps.1001051; 8: 014-020
-
Next Generation Tools in mRNA Purification: The Role of Continuous Raman Spectroscopy Testing with Pretreatment of the SampleLuisetto M*, Nili B Ahmadabadi, Khaled Edbey, Oleg Yurevich Latyshev. Next Generation Tools in mRNA Purification: The Role of Continuous Raman Spectroscopy Testing with Pretreatment of the Sample. Arch Pharm Pharma Sci. 2024: doi: 10.29328/journal.apps.1001052; 8: 021-023
Most Viewed
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic ReviewMohammad Hossein Karami*, Majid Abdouss*, Mandana Karami. Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic Review. Clin J Obstet Gynecol. 2023 doi: 10.29328/journal.cjog.1001151; 6: 201-215
-
Prospective Coronavirus Liver Effects: Available KnowledgeAvishek Mandal*. Prospective Coronavirus Liver Effects: Available Knowledge. Ann Clin Gastroenterol Hepatol. 2023 doi: 10.29328/journal.acgh.1001039; 7: 001-010
-
Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian RandomizationYong-Qing Zhu, Xiao-Yan Meng, Jing-Hua Yang*. Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian Randomization. Arch Asthma Allergy Immunol. 2023 doi: 10.29328/journal.aaai.1001032; 7: 012-022
-
An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergiesNathalie Cottel,Aïcha Dieme,Véronique Orcel,Yannick Chantran,Mélisande Bourgoin-Heck,Jocelyne Just. An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergies. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001027; 5: 030-037

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."