More Information
Submitted: November 21, 2021 | Approved: January 04, 2022 | Published: January 05, 2022
How to cite this article: Mbow M, Diallo I, Diouf M, Cissé M, Gningue M, et al. Evaluation of the LumiraDx SARS-CoV-2 antigen assay for large-scale population testing in Senegal. Int J Clin Virol. 2022; 6: 001-006.
DOI: 10.29328/journal.ijcv.1001041
Copyright License: © 2022 Mbow M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: LumiraDx; SARS-CoV-2; Large-scale testing; RT-PCR; Senegal
Evaluation of the LumiraDx SARS-CoV-2 antigen assay for large-scale population testing in Senegal
Moustapha Mbow1-3*, Ibrahima Diallo1,2*, Mamadou Diouf2*, Marouba Cissé1,2#, Moctar Gningue2#, Aminata Mboup2, Nafissatou Leye2, Gora Lo2,4,5, Yacine Amet Dia2, Abdou Padane2,6, Djibril Wade2, Josephine Khady Badiane2, Oumar Diop7, Aminata Dia2, Ambroise Ahouidi2, Doudou George Massar Niang8, Babacar Mbengue1,8, Maguette Dème Sylla Niang1, Papa Alassane Diaw2, Tandakha Ndiaye Dieye1, Badara Cisé2, El Hadj Mamadou Mbaye2, Alioune Dieye1 and Souleymane Mboup2
1Service d’Immunologie FMPO, Université Cheikh Anta Diop de Dakar, Dakar, Sénégal
2Institut de Recherche en Santé, de Surveillance Épidémiologique et de Formation (IRESSEF), Dakar, Sénégal
3Hôpital Militaire de Ouakam, Dakar, Sénégal
4Centre Médical Interarmées, Dakar, Sénégal
5Service de Bactériologie-Virologie FMPO, Université Cheikh Anta Diop de Dakar, Dakar, Sénégal
6IHU Méditerranée Infection de Marseille Marseille
7Etablissement Public de Santé de Thiès, Sénégal
8Service d’Immunologie du Centre National Hospitalier Universitaire Aristide le Dantec, Dakar, Sénégal
*These equally contributed to this work and share the second authorship
#These equally contributed to this work and share the third authorship
*Address for Correspondence: Dr. Moustapha Mbow, Department of Immunology FMPO, Cheikh Anta Diop University of Dakar, Boulevard Martin Luther King, B.P 5005, Dakar, Sénégal, Email: [email protected]
Purpose: Real-time reverse-transcription polymerase chain reaction (RT-PCR)-based testing remains the gold standard for the diagnosis of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Due to the high diagnosis demand of SARS-CoV-2 and the limited resources for RT-PCR testing, especially in Low-Income Countries (LICs), antigen-based methods are being considered as an option. The aim of this study was to assess the performance of LumiraDx SARS-CoV-2 antigen assay for large population screening compared to RT-PCR.
Methods: This evaluation was conducted on 4146 participants including travelers and participants under household survey and vaccine evaluation studies before injection of the first dose. Oropharyngeal and nasopharyngeal swaps were collected from each participant into 2 mL of viral transport medium (VTM) and 400 μl of VTM were used to assess the performance of LumiraDx SARS-CoV-2 antigen assay, compared to RT-PCR.
Results: The prevalence of SARS-CoV-2 of the cohort was 4.5% with RT-PCR and 4.1% with LumiraDx antigen test. Compared to the RT-PCR, the sensitivity and specificity of the LumiraDx antigen SARS-CoV-2 test were 82,7% [95% CI 74.1-89,7] and 99.9% [95% CI 99.6-99.9] respectively. Given the RT-PCR threshold cycle (Ct) range, the sensitivity was 92.1% [95% CI 84.6-96.3] when the Ct value was below or equal 33 cycles, and 38.1% [95% CI 18.9-61.3] when it was above 33 cycles. The inter-rater reliability showed a kappa coefficient of 0.88 when considering all the patients and 0.94 for Ct values below 33 cycles.
Conclusion: Our data have shown that the LumiraDx platform can be considered for large-scale testing of SARS-CoV-2.
Due to the rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across the world despite the efforts and means spent, identifying better way of isolating infected subjects have become a priority to limit the Coronavirus-related Disease (COVID-19). Rapid and accurate diagnostic tests of SARS-CoV-2 for early confirmation of cases and expediate clinical and public health management decisions may reduce transmission. The current gold standard of SARS-CoV-2 testing relies on the real-time reverse transcription-polymerase chain reaction (RT-PCR) [1,2]. However, some low- and middle-income countries (LMICs) have been struggling to access diagnostic tests due to the large demands in SARS-CoV-2 testing. Indeed, many countries do not have local capacity for manufacturing diagnostic tests and therefore rely on imports. Consequently, with global supplying system, it has become challenging for LMICs to afford pricey equipment or reagents [3]. Beyond the supplying issue, RT-PCR methods appear often time consuming and require special equipment and skilled laboratory personnel. Moreover, PCR testing can lead to longer SARS-CoV2 test results turnaround time which can compromise the ability of people to stay isolated to prevent transmission and spread of the virus [4,5]. For all these reasons, RT-PCR testing may not be suitable for large population testing screening and point-of-care (POC) tests should be considered as frontline testing for SARS-CoV-2 infection diagnosis in order to implement control measures. Most of the antigen rapid diagnostic tests are affordable, simple to perform and allow obtaining results within few minutes [6-13]. Some have the advantage of providing higher throughput for acute SARS-CoV-2 infection, SARS-CoV-2 treatment centres and community-based settings [14]. Despite low sensitivity reported for some SARS-CoV-2 antigen tests, some studies have shown their sensitivity can go up to 93.9% compared to RT-PCR [7,10,11,15-17].
LumiraDx SARS-CoV-2 antigen test (LumiraDx UK Ltd., Dumyat Business Park, Alloa, FK10 2PB, UK), one of the promising tools in targeting the N protein, is a highly conserved target of the virus nucleocapsid, and allows for reliable detection and quantitation. While most of the SARS-CoV-2 test are mainly based on the lateral flow principle, the LumiraDx SARS-CoV-2 antigen test runs on a portable, wall outlet or battery-powered multi-assay point-of-care instrument [18]. The assay reagents are dry single-use, disposable, microfluidic test strips that contain specific antibodies to form an immunoassay complex that uses a fluorescent latex signal to detect the N protein of SARS-CoV-2 [19]. Like most of the POC commercial assays currently available, there are limited data on its performance in large-scale population testing. We have previously assessed LumiraDx SARS-CoV-2 antigen test using oropharyngeal and nasopharyngeal swabs samples collected in viral transport medium (VTM) and found 400 μl as the optimal volume providing high performance of the test (In press). The objective of this study was to evaluate the performance of the LumiraDx platform using VTM samples for large-scale population testing of SARS-CoV-2.
Study populations and samples
We screened 4146 individuals in Dakar, Senegal, including travelers and participants from household survey and vaccine evaluation studies, from March to May 2021. Oropharyngeal and nasopharyngeal swabs were collected from each participant into 2 mL of VTM (KANG JIAN Virus Preservation Medium (Cat: 0201101)) on which RT-PCR test has been performed. Using 400ul of the VTM, which is the volume we previously optimized for the LumiraDx antigen testing (in press), we assessed the performance of LumiraDx Antigen assay for SARS-CoV-2 diagnosis of large population.
This study was approved by the review board of the Senegalese ministry of health (Nr 00000126/MSAS/CNERS/sec). All participants provided informed consent.
Real-time reverse-transcription polymerase chain reaction testing
RNA extraction: The oropharyngeal and nasopharyngeal samples were first inactivated in a water bath at 90 °C for 30 min. The samples were then aliquoted in 1.5 ml vial and RNA was extracted with MagMAX Viral/Pathogen II Nucleic Acid isolation kit using the Kingfisher platform according to the manufacturing and eluted in 50 μl (Thermo Fisher Scientific, Waltham, MA USA, www.thermofisher.com).
Real-time reverse transcriptase-polymerase chain reaction: RNA elution plates were stored at 4 °C while preparing master mix. For SARS-CoV-2 detection, Allplex™ 2019-nCoV assay from Seegene Inc were used according to the manufacturer protocol. Briefly, a master mix of 5 μl of 2019-nCoV MOM, 5 μl of buffer 5 ×, 5 μl of RNase-free water, 1 μl of internal control (IC) and 2 μl of enzymes per sample including negative and positive control were mixed. In each well,
18 μl of master mix were distributed and either 8μl of sample added, 8 μl of positive control or 8 μl of RNase-free water for negative control. Plates were then spun down at 2500 rpm for 5 s and analyzed on a CFX96 Touch Real-Time PCR from BioRad. Reverse Transcription reaction 1 cycle: 50 °C/20 min – 95 °C/15 min. PCR reaction 45 cycles: 94 °C/15 s – 60 °C/30 sec- 72 C/15 sec. Fluorescence was measured at 60° C and 72° C using channels FAM (E gene), HEX (IC), Cal Red 610 (RdRP) and Quasar 670 (N gene). Results were compiled and analyzed using 2019-nCoV viewer from Seegene Inc. according to the manufacturer’s instructions (Seegene. AllplexTM 2019-nCoV Assay) (Cat no. RP10250X/RP10252W) [20]. Results were defined as positive if the viral genome was detected at threshold cycle (Ct) values ≤ 35, as indeterminate at Ct values > 35 and ≤ 38, and as negative at Ct values > 38. Indeterminate results have been excluded from the analysis. We used positive and negative internal controls for quality control.
LumiraDx SARS-CoV-2 antigen testing
The LumiraDx™ SARS-CoV-2 antigen test is a microfluidic immunofluorescence assay for the direct and qualitative detection of nucleocapsid protein antigen of SARS-CoV-2. All the LumiraDx devices underwent lot calibration files for each strip lot, as recommended by the manufacturer, to provide the instruments with information needed to perform diagnostic tests. After oropharyngeal and nasopharyngeal swab sample collection into 2 mL of VTM, 400 μl were added into the extraction vial containing 0.7 ml of extraction buffer. The preparation was inverted gently five times and a drop of the extracted sample was applied onto the sample application area of the inserted test strip. When the sample was detected, the test took 12 min to deliver a positive or negative result. The instrument platform was connected to a cloud server for uploading test data into electronic medical records.
Statistical analysis
To assess the performance of LumiraDx SARS-CoV-2 Antigen test we computed its sensitivity and specificity using the RT-PCR as the gold standard. The agreement between the two methods was evaluated using the Cohen’s kappa coefficient (κ). Statistical analysis was performed using SPSS version 20 (IBM, Inc.) and graphing using GraphPad Prism 5.0 (GraphPad Prism Software Inc., San Diego, California).
From March to mid-June 2021, a total of 4146 oropharyngeal and nasopharyngeal swab samples were collected in VTM from travellers and participants of vaccine evaluation studies. For vaccine study participants, SARS-CoV-2 testing was conducted at enrolment time (day 0 before the first injection). Participants were aged from 2 to 96 years, including 1961 female (47.3%) and 2185 male (52.7%).
Performance of the LumiraDx antigen test for large scale population testing
Using the LumiraDx SARS-CoV-2 antigen assay, the prevalence among the 4146 participants was 4.1% and 4.5% for the RT-PCR. The overall specificity and sensitivity for LumiraDx were 82.7% [95% CI 74.1-89.7] and 99.9% [95% CI 99.6-99.9] respectively compared with the RT-PCR. Considering the Ct threshold value, the LumiraDx SARS-CoV-2 antigen assay was highly sensitive up to and including a threshold cycle (Ct) value of 33 cycles (92.1% [CI 84.6-96.3]). In contrast, its sensitivity was 38.1% [95% CI 18,9-61.3] for Ct values above 33 cycles (Table 1).
| Table 1: Characteristics of the LumiraDx antigen SARS-CoV-2 assay compared to RT-PCR. | ||||
| RT-PCR | LumiraDx Ag Test | |||
| All | All | Ct ≤ 33 | Ct > 33 | |
| Total, n | 4146 | 4146 | 177 | 25 |
| Positive, n(%) | 188(4.5) | 173(4.1) | 152(85,8) | 18(71.4) |
| Negative, n(%) | 3958(95.5) | 3973(95.9) | 25(14.2) | 7(28.6) |
| Sensitivity, % [95% CI] | N/A | 82.7[74.1-89.7] | 92.1[84.6-96.3] | 38.1[18.9-61.3] |
| Specificity, % [95% CI] | N/A | 99.9[99.6-99.9] | 99.9[99,6-99.9] | 100[99.7-100] |
| PPV | N/A | 97.8[91.7-99.6] | 97.9[91,9-99.6] | 100[59.7-100] |
| NPV | N/A | 99.1[98.6-99.5] | 99.9[99.2-99.8] | 94.4[98.9-99.6] |
| Cohen's kappa | N/A | 0.88 | 0.94 | 0.604 |
| Total study population size, the proportion of positive and negative RT-PCR and LumiraDx antigen SARS-CoV-2 results are displayed. The sensitivity, specificity, positive predictive value, negative predictive value and Cohen's kappa of the LumiraDx antigen test compared to RT-PCR are shown for the total study population and ranges of the RT-PCR threshold cycles. Statistical analysis was performed using SPSS version 20 (IBM, Inc.). Abbreviations: PPV: Positive Predictive Value; NPV: Negative Predictive Value; Ct: Threshold Cycle; CI: Confident Interval. | ||||
Analysis of inter-rater agreement between the LumiraDx SARS-CoV-2 antigen assay and the RT-PCR showed a Cohen’s kappa of 0.88, which is almost a perfect agreement [21]. For Ct threshold values up to or equal 33 cycles, kappa was higher with 0.94, whilst it was 0.60 for Ct values above 33 cycles (63.8%) (Table 1).
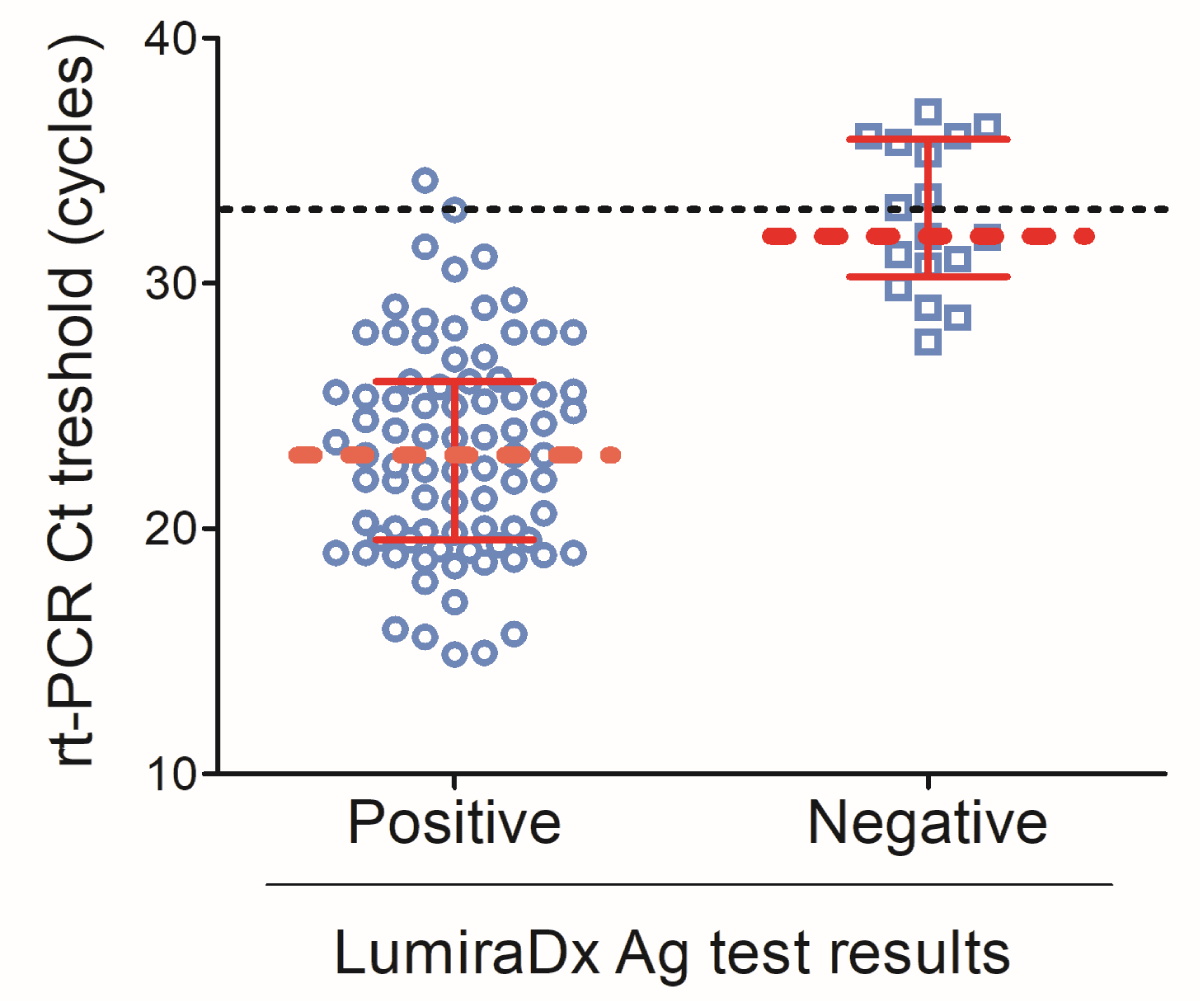
When the RT-PCR threshold cycle was plotted with the LumiraDx antigen results, the median Ct value for LumiraDx positive results was about 22 [IQR: 19.6-26.1] cycles while LumiraDx negative results elicited a median Ct value of about 32 [IQR: 30.3-37.8] (Figure 1).
Figure 1: LumiraDx test results according to the RT-PCR threshold cycle. Scatter plot of the RT-PCR threshold cycle and LumiraDx positive and negative results. Data are shown as median discontinuous line displays the threshold of 33 cycles. Graph has been designed using GraphPad Prism 5.0.
Because of the emergence of SARS-CoV-2 variants and the possible seasonality of the virus, it is crucial to have large capacity of testing for early implementation of control measures. Due to several limitations in RT-PCR testing capacity, especially in LMIC, the development of point-of-care tests as frontline testing for SARS-CoV-2 is crucial to proceed to large population testing. Several SARS-CoV-2 antigen tests have been developed and are now available on the market [7-13]. Such emerging antigen diagnosis tools, although affordable and easy-to-use, have limited data on their performance. Here, we evaluate the LumiraDx SARS-CoV-2 antigen test, an immunofluorescence-based assay targeting the highly conserved N protein of SARS-CoV-2, for large-scale population testing of SARS-COV-2 in community settings. In our knowledge, this is the first study evaluating the performance of the LumiraDx antigen SARS-CoV-2.
Using oropharyngeal and nasopharyngeal swabs samples from individuals of the general population and participants of vaccine evaluation studies, the prevalence of SARS-CoV-2 using the RT-PCR testing was 4.5% and 4.1% with LumiraDx antigen test. This evaluation was conducted between March and May 2021 when little cases were reported in Senegal [22,23], which may explain the low prevalence of SARS-CoV-2 in this cohort. When compared to RT-PCR, the LumiraDx SARS-CoV-2 antigen assay displayed an overall sensitivity of 82.7% and specificity of 99.9%. With a kappa of 0.88, the agreement between RT-PCR and LumiraDx was almost perfect [21] when considering whole study population. Such sensitivity is lower than what the LumiraDx company reported [24] although we found greater specificity. Such difference in the specificity we found is in line with several studies showing lower performance of many antigen tests to what the manufacturers reported [7,10,11,13,15-17]. Moreover, the company performance has been evaluated in patients whose period of symptoms occurred within 12 days since onset while we did not report symptom of our study participants. Nevertheless, the sensitivity found in our study meets the WHO performance requirements for the use of SARS-CoV-2 Ag-RDTs of ≥ 80% sensitivity and ≥ 97% specificity when compared to RT-PCR a reference assay [25]. Interestingly, the sensitivity of the LumiraDx SARS-CoV-2 antigen assay was found to be function of the RT-PCR threshold cycle as certified by the sensitivity was 92.1% for Ct values below or equal 33 cycles whilst above 33 cycles it was 38.1%. This is reinforced but the study of Drain, et al. also shows that higher sensitivity for Ct values below 33 cycles [18] and highlights the need for a high-sensitivity test to identify individuals with a low viral load.
Whether the low sensitivity of LumiraDx for RT-PCR Ct value above 33 cycles may impede its use in control strategies of the COVID-19 pandemic need to be determined before its implementation in any SARS-CoV-2 testing guideline. Several studies have shown that lower Ct values from respiratory samples was associated with more severe disease [26-35] but the key question is how the high Ct values elicited in non-severe diseases impacts on the transmission. Investigations on the pathogenicity of the virus have shown that its infectivity, defined as growth in cell culture, is associated to high viral load (low RT-PCR Ct value) [36-38]. Indeed, Ct values below 33 cycles have shown to be associated with higher probability of a positive viral culture [36] and Ct value increase from day 10 since symptom onset [39]. Impact on positivity of virus culture has also been found to be significantly associated to decrease in Ct by demonstrating that for every unit increase in Ct, the odds of positive culture decreased by 32% [40]. Overall, this and the relatively high Ct values we observed in individuals with false-negative antigen testing results compared to RT-PCR, as also previously reported in many studies [7-13,16], may suggest that from 33 cycles, positive RT-PCR tests may reveal noncontagious remnant viral RNA.
A limitation of this was that clinical characteristics were not reported, which did not allow evaluation of the performance according to the time the onset of the symptom.
We found here that RT-PCR and the LumiraDx SARS-CoV-2 antigen test display a very similar prevalence for large SARS-CoV-2 testing at community level. The LumiraDx test displayed sensitivity that meets the WHO performance requirements for the use of SARS-CoV-2 antigen testing compared to RT-PCT. Its sensitivity was even highly improved for RT-PCR Ct values up to 33 and from which positive RT-PCR results are considered by some authors as noncontagious remnant viral RNA. Overall, our data shows that the LumiraDx SARS-CoV-2 platform might be suitable for large-scale population testing.
Funding
This study was funded by the “West African Task Force for the Control of Emerging and Re-emerging Infectious Diseases” (WATER), and “Innovation in Laboratory Engineered Accelerated Diagnosis” (iLEAD) (Grant Nr. OPP1214434/INV-009631). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed are those of the authors, and the funders are not responsible for any use that may be made of the information contained herein.
Availability of data and materials: The data that support the findings of this study are available upon request from the authors.
Code availability: Statistical analysis was performed using SPSS version 20 (IBM, Inc.) and graphing using GraphPad Prism 5.0 (GraphPad Prism Software Inc., San Diego, California).
Ethics approval: This study was approved by the “Comité National d’Ethique pour la recherche en Santé” of the Senegalese Ministry of Health (Authorisation Nr: 00000110/MSAS/CNERS/Sec).
Consent to participate: Informed and written consent were obtained from all participants; for children, consent was obtained from their parent or a legal guardian.
Consent for publication: All authors have seen and approved the content of the manuscript and have contributed significantly to the work
Authors’ contributions: MM, SM, AD conceived and designed the study. ID, MG, MC, MD, NL and GL performed the experiments. MM, ID, MG, MC, MD, GL, and NL recruited study participants and collected data. MM analyzed and interpreted the data. SM, AD, MM, EMM, DW, NL, GL contributed to reagents/materials/analysis tools. MM, ID, MG, MC, MD, AD, GL, NL participated to study design. MM, SB, participated to study coordination. MM, GL wrote/drafted the manuscript. YAD, DW, NL, GL, JKB, AP, OD, AD, AA, MN, DGN, PAD, AM, BM, MDSN, TND, EMM reviewed critically the manuscript for important intellectual content. SM and AD approved the final version to be published. All authors approved the final version of the manuscript.
We gratefully thank the individuals participating in this study (travelers and volunteers of vaccine evaluation studies), the staff of IRESSEF involved in the SARS-CoV-2 testing. We also thank Maria Yazdanbakhsh, head of the Parasitology and Cellular Immunology Department of Leiden University Medical Center, and her team for their support in reagents and consumables supply.
- Mathuria JP, Yadav R, Rajkumar. Laboratory diagnosis of SARS-CoV-2 - A review of current methods. J Infect Public Health. 2020; 13: 901-905. PubMed: https://pubmed.ncbi.nlm.nih.gov/32534946/
- WHO. Interim Guidance; 2020. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. CDC C-NC-nR-TR-PDP CDC. 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.
- Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020; 38: 515-518. PubMed: https://pubmed.ncbi.nlm.nih.gov/32203294/
- Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004; 10: 311-316. PubMed: https://pubmed.ncbi.nlm.nih.gov/15030703/
- Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, et al. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano. 2020; 14: 3822-3835. PubMed: https://pubmed.ncbi.nlm.nih.gov/32223179/
- Farfour E, Asso-Bonnet M, Vasse M. The ID NOW COVID-19, a high-speed high-performance assay. Eur J Clin Microbiol Infect Dis. 2021; 40: 2041-2045. PubMed: https://pubmed.ncbi.nlm.nih.gov/33855651/
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020; 99: 328-333. PubMed: https://pubmed.ncbi.nlm.nih.gov/32497809/
- Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol. 2020; 58: e01438-01420. PubMed: https://pubmed.ncbi.nlm.nih.gov/32636214/
- Krogh J. Empyema of the gallbladder: a case with unusual presentation. Acta Chir Belg. 1989; 89: 204-205. PubMed: https://pubmed.ncbi.nlm.nih.gov/2800855/
- Linares M, Perez-Tanoira R, Carrero A, Romanyk J, Perez-Garcia F, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020; 133: 104659. PubMed: https://pubmed.ncbi.nlm.nih.gov/33160179/
- Mertens P, De VN, Martiny D, Jassoy C, Mirazimi A, et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front Med (Lausanne). 2020; 7: 225. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7227790/
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, et al. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J Clin Med. 2021; 10: 328. PubMed: https://pubmed.ncbi.nlm.nih.gov/33477365/
- Mak GCK, Lau SSY, Wong KKY, Chow NLS, Lau CS, et al. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J Clin Virol. 2021; 134: 104712. PubMed: https://pubmed.ncbi.nlm.nih.gov/33338894/
- World Health Organization. A coordinated global research roadmap: 2019 novel coronavirus. 2020. https://www.who.int/blueprint/priority-diseases/key-action/Coronavirus_Roadmap_V9.pdf
- Nalumansi A, Lutalo T, Kayiwa J, Watera C, Balinandi S, et al. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis. 2021; 104: 282-286. PubMed: https://pubmed.ncbi.nlm.nih.gov/33130198/
- Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, et al. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020; 129: 104455. PubMed: https://pubmed.ncbi.nlm.nih.gov/32485618/
- Toptan T, Eckermann L, Pfeiffer AE, Hoehl S, Ciesek S, Drosten C, et al. Evaluation of a SARS-CoV-2 rapid antigen test: Potential to help reduce community spread? J Clin Virol. 2021; 135: 104713. PubMed: https://pubmed.ncbi.nlm.nih.gov/33352470/
- Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, et al. A Rapid, High-Sensitivity SARS-CoV-2 Nucleocapsid Immunoassay to Aid Diagnosis of Acute COVID-19 at the Point of Care: A Clinical Performance Study. Infect Dis Ther. 2021; 10: 753-761. https://pubmed.ncbi.nlm.nih.gov/33629225/
- LumiraDx. LumiraDx website and SARS-CoV-2 Antigen test EUA Product Insert. 2020. https://www.lumiradx.com/us-en/
- Instructions for Use. https://www.fda.gov/media/137178/download
- McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012; 22: 276-282. https://pubmed.ncbi.nlm.nih.gov/23092060/
- Woldometer. https://www.worldometers.info/coronavirus/country/senegal
- Ministère de la Sante et de l’Action Sociale. https://www.sante.gouv.sn
- LumiraDx. Performance evaluation of the LumiraDx SARS-CoV-2 Antigen Test to aid diagnosis of acute COVID-19 at the point of care. https://www.lumiradx.com/assets/pdfs/white-papers/performance-evaluation-of-sars-cov-2-ag-test.pdf?v=1
- WHO Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance. 2020. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays
- Huang JT, Ran RX, Lv ZH, Feng LN, Ran CY, et al. Chronological Changes of Viral Shedding in Adult Inpatients With COVID-19 in Wuhan, China. Clin Infect Dis. 2020; 71: 2158-2166. PubMed: https://pubmed.ncbi.nlm.nih.gov/32445580/
- Liu Y, Yan LM, Wan L, Xiang TX, Le A, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020; 20: 656-657. PubMed: https://pubmed.ncbi.nlm.nih.gov/32199493/
- Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation Between Relative Nasopharyngeal Virus RNA Load and Lymphocyte Count Disease Severity in Patients with COVID-19. Viral Immunol. 2021; 34: 330-335. PubMed: https://pubmed.ncbi.nlm.nih.gov/32297828/
- Liu Y, Yang Y, Zhang C, Huang F, Wang F, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020; 63: 364-374. PubMed: https://pubmed.ncbi.nlm.nih.gov/32048163/
- Xia XY, Wu J, Liu HL, Xia H, Jia B, et al. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J Clin Virol. 2020; 127: 104360. PubMed: https://pubmed.ncbi.nlm.nih.gov/32305025/
- Yu X, Sun S, Shi Y, Wang H, Zhao R, et al. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care. 2020; 24: 170. PubMed: https://pubmed.ncbi.nlm.nih.gov/32326952/
- Zou L, Ruan F, Huang M, Liang L, Huang H, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020; 382: 1177-11779. PubMed: https://pubmed.ncbi.nlm.nih.gov/32074444/
- Schwierzeck V, Konig JC, Kuhn J, Mellmann A, Correa-Martinez CL, et al. First Reported Nosocomial Outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 in a Pediatric Dialysis Unit. Clin Infect Dis. 2021; 72: 265-270. PubMed: https://pubmed.ncbi.nlm.nih.gov/33501962/
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med. 2020; 382: 2081-2090.
- Kimball A, Hatfield KM, Arons M, James A, Taylor J, et al. Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility - King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020; 69: 377-381. PubMed: https://pubmed.ncbi.nlm.nih.gov/32240128/
- La SB, Le BM, Andreani J, Hoang VT, Grimaldier C, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020; 39: 1059-1061. PubMed: https://pubmed.ncbi.nlm.nih.gov/32342252/
- Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, et al. SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerg Infect Dis. 2020; 26: 2701-2704. PubMed: https://pubmed.ncbi.nlm.nih.gov/32749957/
- van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun. 2021; 12: 267. PubMed: https://pubmed.ncbi.nlm.nih.gov/33431879/
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020; 581: 465-469. PubMed: https://pubmed.ncbi.nlm.nih.gov/32235945/
- Bullard J, Dust K, Funk D, Strong JE, Alexander D, et al. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin Infect Dis. 2020; 71: 2663-2666. PubMed: https://pubmed.ncbi.nlm.nih.gov/32442256/